Back to top
Conservation Genetic Methods

Genetic methods provide new insights into the biology of species as well as the spatial and temporal structure of populations and will therefore challenge nature conservation management.
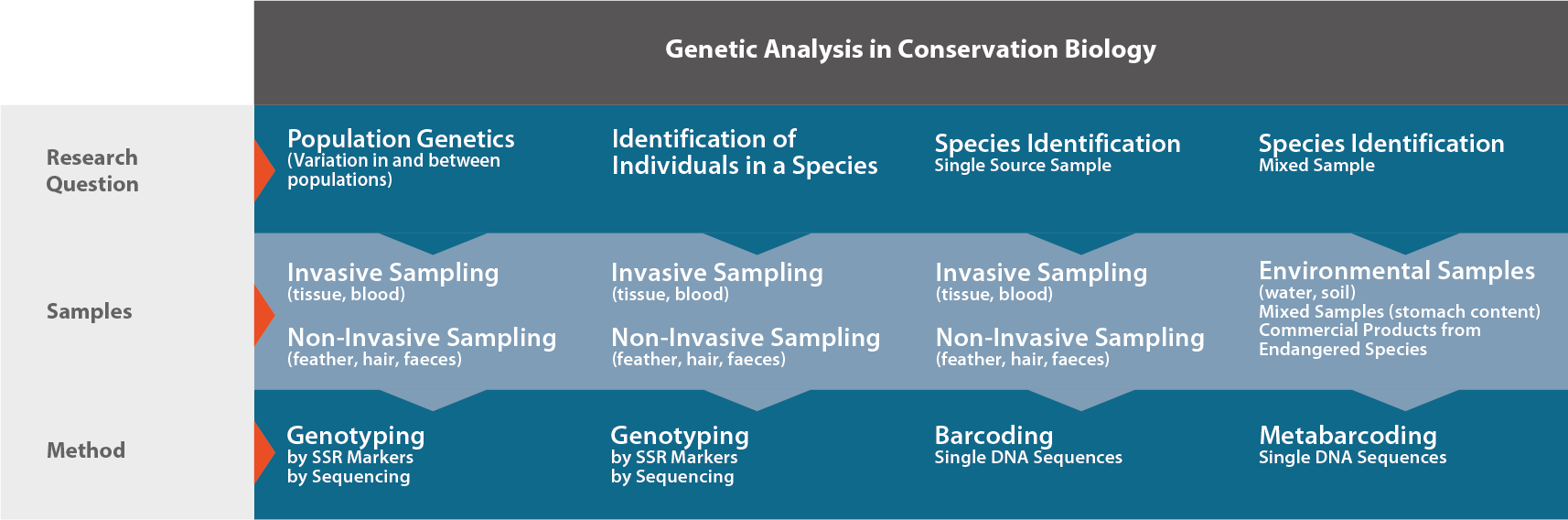
Any conservation genetics study is initiated by research questions defined by experts in nature conservation management. Depending on the question, conservation genetics provide a broad variety of methodologies which will in turn define the experimental layout, the statistical analysis of the data and the recommendations for practice. All these steps such as sample collection, genetic laboratory analyses, statistical evaluations, interpretation and conclusions, must be carefully planned for each project. Focus on the practical aspects in nature conservation and let Ecogenics run the complete laboratory part including marker development, if needed, and the genotyping of samples. Moreover, we can help you define the sampling scheme for your study and help analyze and interpret the data derived from the study.
Overview
Methods
Genotyping

Biodiversity is built on three levels. The basis is genetic diversity, followed by the diversity of species and then the diversity of ecosystems. By genotyping individuals of a single species collected from different populations using appropriate genetic markers, insights into the genetic diversity are possible, allowing one to draw conclusions about the behaviour of the species.
The most commonly used markers to study genetic variation are microsatellite markers (SSR: simple sequence repeat). Microsatellite markers are species specific, inherited from both parents (co-dominant inheritance), selectively neutral and normally highly polymorphic. By genotyping a set of different microsatellite markers, each individual will show a distinct allelic pattern allowing one to identify individuals and to determine the population size.
Moreover, the frequencies of alleles among individuals collected from different populations can be used to calculate population genetic measures such as the genetic variation in and between populations and draw conclusions on the populations genetic structure, inbreeding, hybridization and gene flow. Gene flow is a measure of genetic exchange between populations and allows one to answer the question on the cross-linking of populations. In addition, by analysing the population structure at different time points, assessment of the extent of genetic diversity and its change over time can be made. The analysis of the population structure before and after conservation management measures to reduce habitat fragmentation can, for example, be studied for specific species to see whether the taken measures were successful.
Small amounts of DNA are already sufficient to perform a genotyping by microsatellite analysis and ecogenics has expertise in DNA isolation from non-invasive samples (faeces, hair, feathers, egg shell, etc.). This method is also suitable for various species that are difficult to observe. For example, the number of individuals in endangered species such as Tetrao urogallus (Capercaille) can be estimated without invasive sampling and the resulting data can be used for parentage analysis.
In order to ensure highest data quality and to speed up the progress of your projects, ecogenics offers the full range of genotyping services. Hence, we are happy to carry out your genotyping projects, based on markers developed by ecogenics, or on markers from other sources (in-house, published etc.). Our vast experience with DNA isolation from many sources, combined with our expertise in PCR and fragment length analysis, ensures optimal results. Whether you wish to receive fluorescent trace files (.fsa), an allele table with all identified alleles per marker and sample, or further bioinformatics information, is up to you to decide.
Please don’t hesitate to contact us. We are happy to discuss your planned project and provide you with an individually tailored quote.
Barcoding

Classification of species is often hampered by the fact that the structure for a morphological classification such as reproductive structures in plants or fungi are not present. Moreover, many species are difficult to classify due to minute morphological differences between species which require an in-depth expert knowledge. DNA based barcoding can help to overcome some of the restrictions of the classical classification of species. The research community invested considerable effort to find DNA-based markers for molecular species classification. The genetic identification of a species is based on the analysis of a short DNA sequence (mainly from mitochondria and chloroplasts or ribosomal DNA) which can be compared to a reference database.
ecogenics offers a full service from DNA isolation to species identification for all groups of organisms. Our quality service includes DNA isolation from many sources (tissue, blood, hair, faeces, feather, egg shell, etc.) followed by PCR amplification of the appropriate barcoding locus for the respective organismal group (e.g. 16S, ITS, mitochondrial Cytochrome b, Cytochrome c Oxidase I, control region) and Sanger sequencing. Finally, we identify the species based on publicly available databases and provide you with a species identification certificate.
Metabarcoding

Metabarcoding is a rapid method of biodiversity assessment that combines the use of DNA based identification (barcoding) and high-throughput DNA sequencing. This allows one to assess the presence of communities within a sample from minute amounts of DNA isolated from environmental samples such as stool, water or soil. A classical application of metabarcoding is the study of bacterial and fungal communities. But the same approach can also be applied to nearly any organismal group, e.g. the detection of amphibian DNA in water samples or animal diet analysis.
Depending on the aim of the analysis, a critical issue in metabarcoding analysis is the availability of a well curated reference database to link the sequences generated during the analysis of the samples to the taxonomy. Although many public reference databases are available for some organismal groups, in some cases it may also be beneficial to set-up a proprietary reference database using well characterized specimens.
Ecogenics developed and validated different metabarcoding applications and optimized the bioinformatic analysis to analyze the next generation’s sequencing data. Our quality service includes DNA isolation from many sources (water, stool, soil, etc.) followed by PCR amplification of the appropriate barcoding locus for metabarcoding using universal primers, next-generation sequencing as well as bioinformatics analysis.
Marker Systems
Available Marker Systems for:
-
Genotyping Indicator Species
Connecting biotopes is an important nature conservation strategy in Switzerland. Indicators for connectivity and/or fragmentation of biotopes are widespread, medium-abundant species with a leading species function for habitats. Ecogenics offers molecular genetic analysis for the following indicator species:
Melittis melissophyllum (Immenblatt)
Stethophyma grossum (Sumpfschrecke)
Metrioptera bicolor (= Bicolorana bicolor) Zweifarbige Beisschrecke
Zebrina detrita (Zebraschnecke, Weisse Turmschnecke)
Bombina variegata variegata (Gelbbauchunke)
Triturus cristatus & Triturus carnifex
- Metabarcoding: Amphibian detection
Amphibians have a particularly high priority in both biotope and species conservation. For the monitoring of amphibians in aquatic ecosystems, ecogenics offers amphibian detection and identification based on molecular genetic analysis by eDNA (water sample) metabarcoding.
- Barcoding: BOLDSYSTEMS
Our molecular genetic analyses with universal marker systems allows species identification with the Barcode of Life Data System (bold), an informatics workbench aiding the acquisition and publication of DNA barcode records.
Animal Identification (COI)
Fungal Identification (ITS)
Plant Identification (RBCL & MATK)
If you have not yet found a suitable marker system for your species or research question, please do not hesitate to contact us. Marker systems have already been developed for many species and details can often be found in the literature. Besides, ecogenics is specialized in the development of marker systems and we would be happy to work with you on your project. Get in contact with us to see how we can enrich your research.