Back to top
Genotyping by SSRs

In order to ensure the highest data quality and to speed up the progress of your research projects, ecogenics offers the full range of genotyping services. Hence, we are happy to carry out your genotyping projects, based on markers developed by ecogenics, or on markers from other sources (in-house, published etc.). Our vast experience with DNA isolation from many sources, combined with our expertise in PCR ensures optimal results in a short time (6 to 10 weeks, depending on the project size).
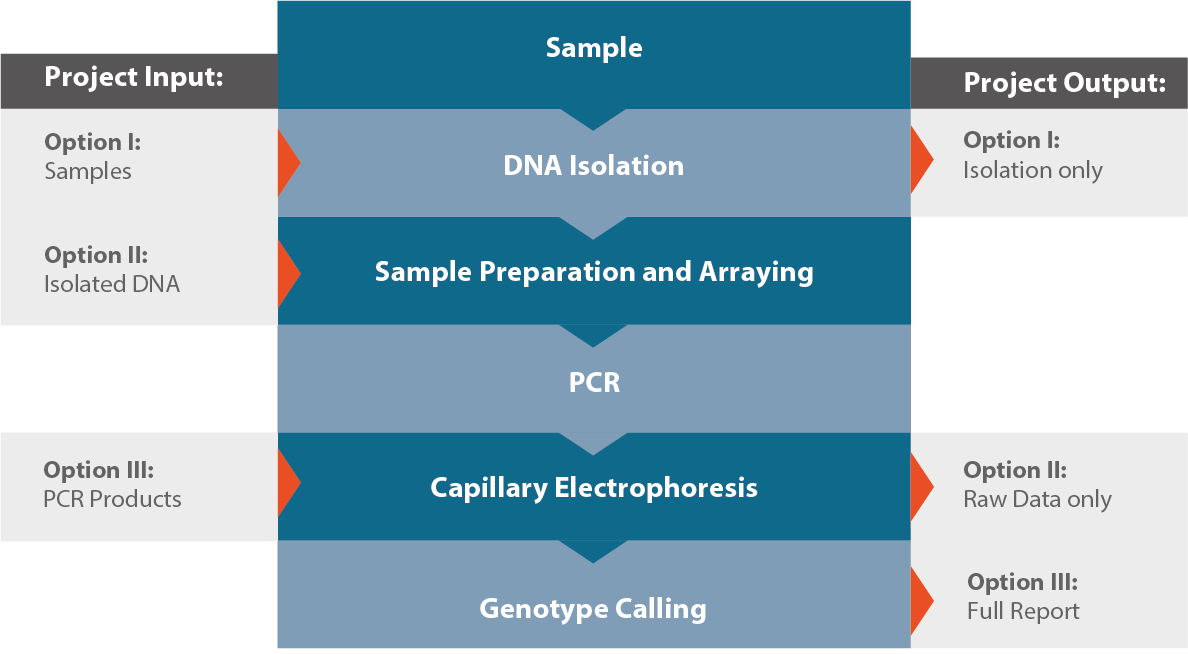
Workflow
Products
SSR Multiplexing
Multiplex PCR reactions are more cost-effective to run compared to singleplex reactions for genotyping projects involving larger numbers of samples and markers. Therefore, ecogenics provides a service for the Multiplexing of SSR markers and depending on the number of markers and samples, we recommend using 3 to 7-plex assays. The SSR markers can either result from an SSR development project run at ecogenics or any other published SRR markers.
By default, we use the fluorescent dyes FAM, ATTO532, ATTO550 and ATTO565, compatible with ABI’s standard filter set G5. Besides detailed assay description, customers receive 100 µM stock solutions of all primers used in the assay, including dye-labelled forward primers.
Published SSR Marker Testing
In case microsatellite markers have been previously developed for your species of interest and you are not sure whether these published markers perform stably on your species/subspecies we will test functionality and polymorphy for each locus assayed in 7 individuals.
Besides detailed assay description, customers receive 100 µM stock solutions of the primers of each locus tested, with the forward primers carrying a FAM label.
PCR & Capillary Electrophoresis
ecogenics has a strong track record in PCR and capillary electrophoresis. Our processes are optimized and automated with high standards resulting in a low error rate and reliable data.We use the Applied Biosystems 3730xl DNA Analyzer platform for capillary electrophoresis. By default, we use the fluorescent dyes FAM, ATTO532, ATTO550 and ATTO565, compatible with ABI’s standard filter set G5.
Whether you wish to receive fluorescent trace files (.fsa) or also an allele table with all identified alleles per marker and sample is up to you to decide.
Genotype Calling
With our analysis service, fluorescent trace files (.fsa) are subjected to automated analysis procedures with the analysis tool SoftGenetics GeneMarker®. Once the analysis with GeneMarker® has been completed, the validity of each allele call is verified by visual inspection. This guarantees a high accuracy of the results provided. The delivery includes an allele table (.xlsx) with all identified alleles per marker and sample.
Results
The delivery of SSR genotyping project includes:
-
a genotyping project summary
-
the electropherograms as raw data in an ABI-Format (.fsa)
-
stock solutions of the labelled and unlabelled primers (100 µM)
-
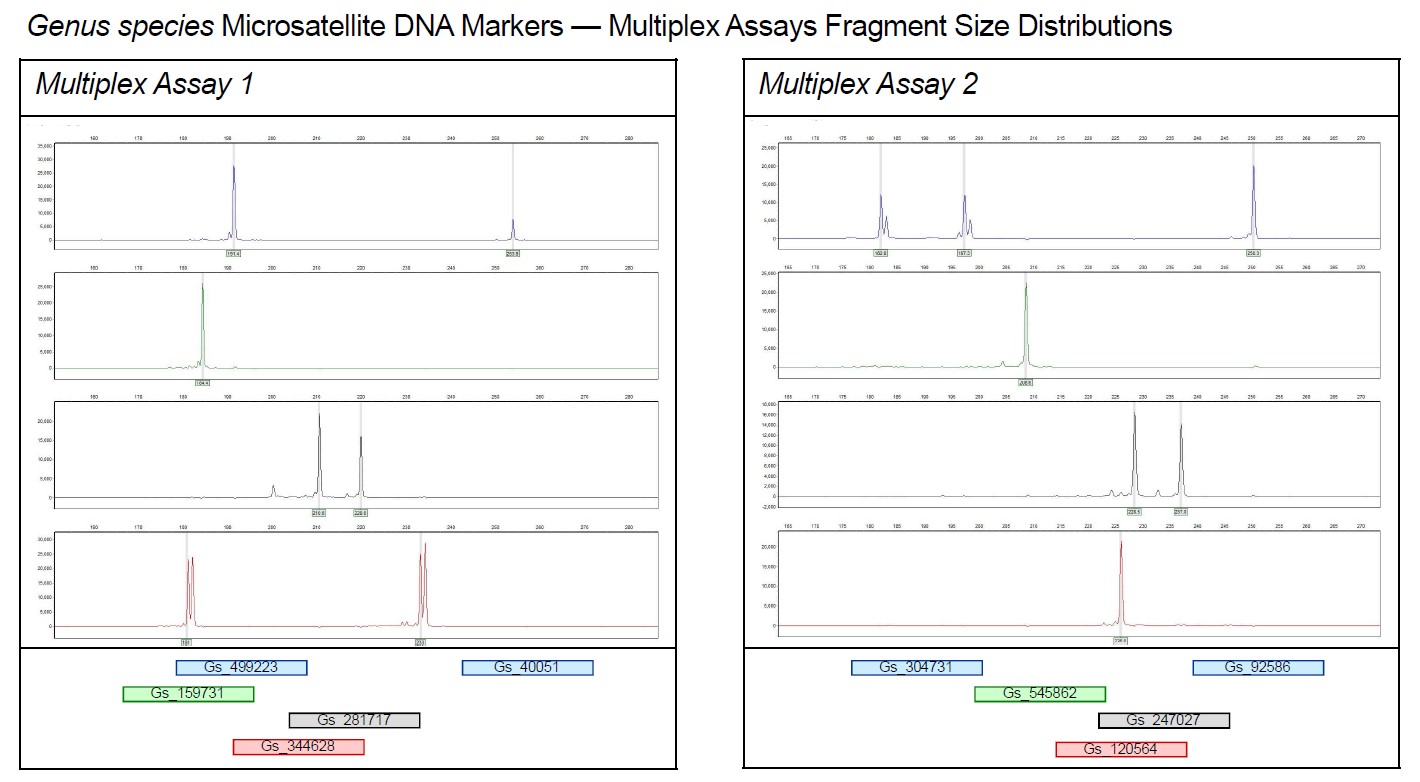
If a SSR Multiplexing has been run before the genotyping, a Read&Go brochure containing detailed information on the multiplexed PCR, the corresponding PCR protocols and a picture of the electropherogram (see Figure 1) is part of the delivery.
-
If you have additionally chosen the product Genotype Calling, an excel file (see Figure 2) containing the allele sizes per locus and sample are part of the delivery.